factors_r = ["SP500", "DTWEXAFEGS"] # "SP500" does not contain dividends; note: "DTWEXM" discontinued as of Jan 2020
factors_d = ["DGS10", "BAMLH0A0HYM2"]Decomposition
Underlying returns are structural bets that can be analyzed through dimension reduction techniques such as principal components analysis (PCA). Most empirical studies apply PCA to a covariance matrix (note: for multi-asset portfolios, use the correlation matrix because asset-class variances are on different scales) of equity returns (yield changes) and find that movements in the equity markets (yield curve) can be explained by a subset of principal components. For example, the yield curve can be decomposed in terms of shift, twist, and butterfly, respectively.
\[ \begin{aligned} \boldsymbol{\Sigma}&=\lambda_{1}\mathbf{v}_{1}\mathbf{v}_{1}^\mathrm{T}+\lambda_{2}\mathbf{v}_{2}\mathbf{v}_{2}^\mathrm{T}+\cdots+\lambda_{k}\mathbf{v}_{k}\mathbf{v}_{k}^\mathrm{T}\\ &=V\Lambda V^{\mathrm{T}} \end{aligned} \]
def eigen(x):
L, V = np.linalg.eig(np.cov(x.T, ddof = 1))
idx = L.argsort()[::-1]
L = L[idx]
V = V[:, idx]
result = {
"values": L,
"vectors": V
}
return resultdef eigen_decomp(x, comps):
LV = eigen(x)
L = LV["values"][:comps]
V = LV["vectors"][:, :comps]
result = np.dot(V, np.multiply(L, V.T))
return np.ravel(result)comps = 1eigen_decomp(overlap_df, comps) * scale["periods"] * scale["overlap"]array([ 3.21061014e-02, -3.84353444e-03, -7.78728776e-05, 2.64410445e-03,
-3.84353444e-03, 4.60123041e-04, 9.32243637e-06, -3.16535054e-04,
-7.78728776e-05, 9.32243637e-06, 1.88879522e-07, -6.41323654e-06,
2.64410445e-03, -3.16535054e-04, -6.41323654e-06, 2.17755755e-04])# np.cov(overlap_df.T) * scale["periods"] * scale["overlap"]Variance
We often look at the proportion of variance explained by the first \(i\) principal components as an indication of how many components are needed.
\[ \begin{aligned} \frac{\sum_{j=1}^{i}{\lambda_{j}}}{\sum_{j=1}^{k}{\lambda_{j}}} \end{aligned} \]
def variance_explained(x):
LV = eigen(x)
L = LV["values"]
result = L.cumsum() / L.sum()
return resultvariance_explained(overlap_df)array([0.88636218, 0.99272118, 0.9982564 , 1. ])Similarity
Also, a challenge of rolling PCA is to try to match the eigenvectors: may need to change the sign and order.
\[ \begin{aligned} \text{similarity}=\frac{\mathbf{v}_{t}\cdot\mathbf{v}_{t-1}}{\|\mathbf{v}_{t}\|\|\mathbf{v}_{t-1}\|} \end{aligned} \]
def similarity(V, V0):
n_cols_v = V.shape[1]
n_cols_v0 = V0.shape[1]
result = np.zeros((n_cols_v, n_cols_v0))
for i in range(n_cols_v):
for j in range(n_cols_v0):
result[i, j] = np.dot(V[:, i], V0[:, j]) / \
np.sqrt(np.dot(V[:, i], V[:, i]) * np.dot(V0[:, j], V0[:, j]))
return resultdef roll_eigen1(x, width, comp):
n_rows = len(x)
result_ls = []
for i in range(width - 1, n_rows):
idx = range(max(i - width + 1, 0), i + 1)
LV = eigen(x.iloc[idx])
V = LV["vectors"]
result_ls.append(V[:, comp - 1])
result_df = pd.DataFrame(result_ls, index = x.index[(width - 1):],
columns = x.columns)
return result_df comp = 1raw_df = roll_eigen1(overlap_df, width, comp)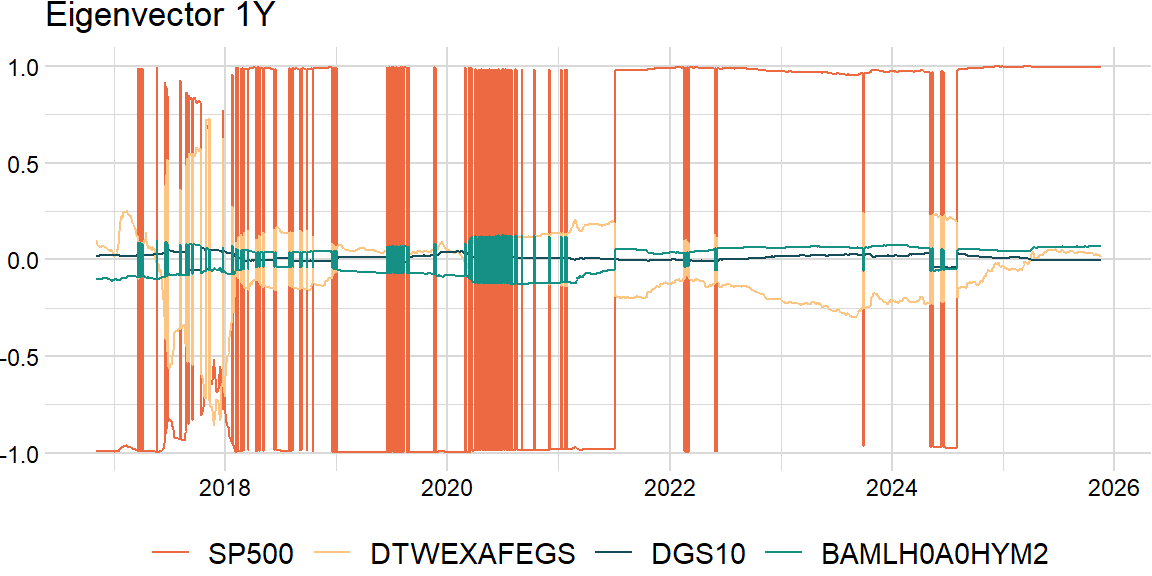
def roll_eigen2(x, width, comp):
n_rows = len(x)
V_ls = []
result_ls = []
for i in range(width - 1, n_rows):
idx = range(max(i - width + 1, 0), i + 1)
LV = eigen(x.iloc[idx])
V = LV["vectors"]
if i > width - 1:
# cosine = np.dot(V.T, V_ls[-1])
cosine = similarity(V.T, V_ls[-1])
order = np.argmax(np.abs(cosine), axis = 1)
V = np.sign(np.diag(cosine[:, order])) * V[:, order]
V_ls.append(V)
result_ls.append(V[:, comp - 1])
result_df = pd.DataFrame(result_ls, index = x.index[(width - 1):],
columns = x.columns)
return result_dfclean_df = roll_eigen2(overlap_df, width, comp)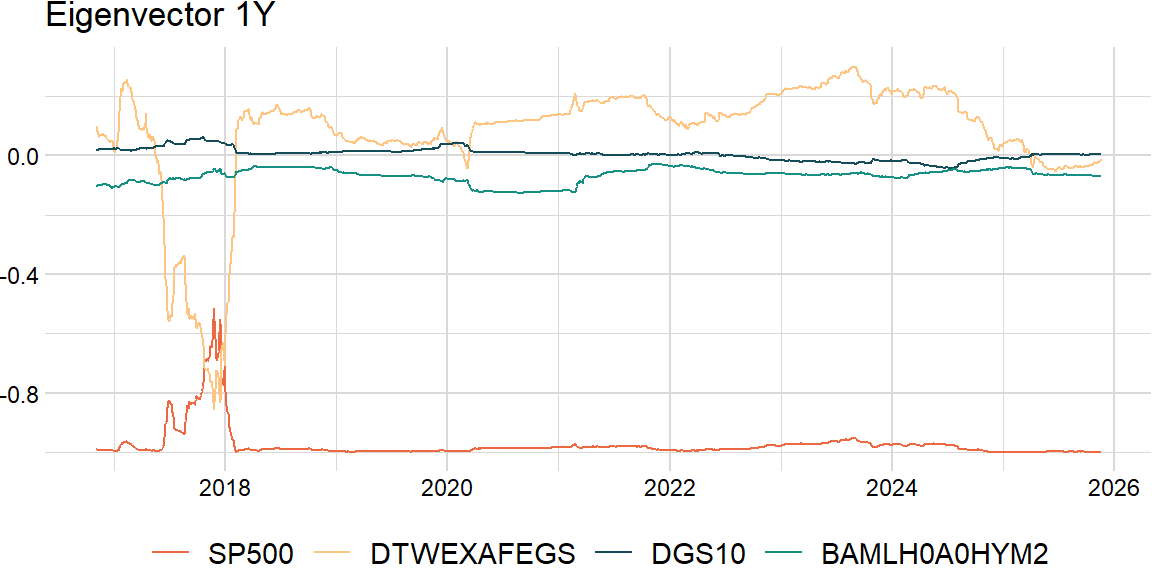
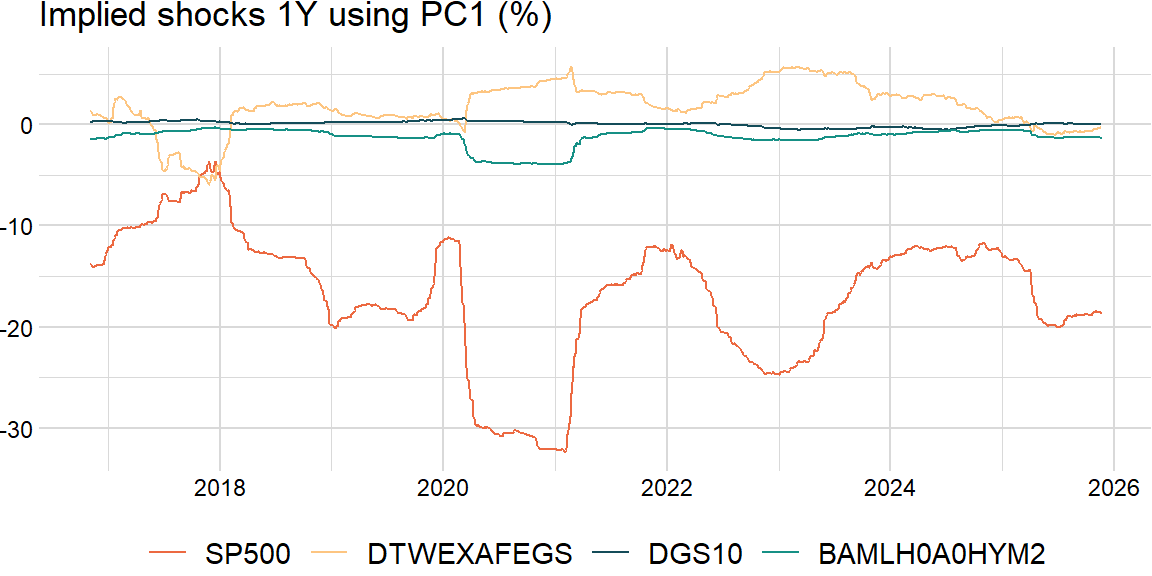
Implied shocks
Product of the \(n\)th eigenvector and square root of the \(n\)th eigenvalue:
def roll_shocks(x, width, comp):
n_rows = len(x)
V_ls = []
result_ls = []
for i in range(width - 1, n_rows):
idx = range(max(i - width + 1, 0), i + 1)
LV = eigen(x.iloc[idx])
L = LV["values"]
V = LV["vectors"]
if len(V_ls) > 1:
# cosine = np.dot(V.T, V_ls[-1])
cosine = similarity(V.T, V_ls[-1])
order = np.argmax(np.abs(cosine), axis = 1)
L = L[order]
V = np.sign(np.diag(cosine[:, order])) * V[:, order]
shocks = np.sqrt(L[comp - 1]) * V[:, comp - 1]
V_ls.append(V)
result_ls.append(shocks)
result_df = pd.DataFrame(result_ls, index = x.index[(width - 1):],
columns = x.columns)
return result_dfshocks_df = roll_shocks(overlap_df, width, comp) * np.sqrt(scale["periods"] * scale["overlap"])